GIS can change with current technology such as Apache Arrow, and perhaps also with some other techniques. In Pythonland, Geopandas has improved performance over time. I also wanted to try Dask, DuckDB and Apache Sedona.
Please contact me if you read this and think I could have improved the code quality and speed. I use some public data, which is still widely used in Germany as shape files.
Update: In the previous version I was not able to save to geoparquet with DuckDB. It works, it can be openmed inm QGIS, not so far no CRS.
The data
First I downloaded the ALKIS (register) building data for all counties in the state of Brandenburg. All the vector files are open data. The vector files are still offered as shapefiles. From the ALKIS dataset of Brandenburg I used the buildings and the parcels (with land use). The files are stored per county! The geometries have some errors, which Geopandas automatically detects and fixes. In addition, some files cannot be opened with the fiona library of Geopandas, with the error message of multiple geometry columns. So we always use the new default: pyogrio.
Task
- Open the datasets and concatenate the counties.
- Create an intersection overlay
- Save the data, if possible as a geoparquet file.
Why did I choose this task? I think the overlay is one of the more computationally intensive tasks in GIS. I may write articles about other tasks later. In Geopandas uses a spatial index, then calculates an intersection and joins the original data to the intersection.
We save it as a geoparquet file, because that’s the only format Dask-GeoPandas can write to. In addition, the result (with good compression) is small (391 MB) compared to what Geopackage (1.57 GB) needs.
By the way, I choose which columns to open in Geopandas, because I will find out later that one column contains only none.
So I just don’t use unimportant columns from the beginning.
The hardware
I ran the speed test on a WIN10 PV with a Ryzen 7 5800X with 48 GB of RAM. The final runs are done with Hyperfine and 3 warm-up runs and the default 10 runs, 5 for DuckDB1.
The memory usage tests are done on a laptop with Ryzen 7 4800 and 32 GB of RAM running TuxedoOS. The reason for this is that RAM usage is only fully recorded under Linux.
The Frameworks
Geopandas
For me, Geopandas has been the goto solution for years. Sometimes with some extra code, some extra libs like pyogrio.
Expectations: Well, nothing special. It just works. Should load faster with pyogrio.
Observations: Initially, loading the data takes about 75 to 80 seconds on my machine with an AMD Ryzen 5800X CPU.
It’s a bit faster when using arrow by about 15 seconds.
It got a bit slower when dropping the duplicates (on the district borders) by there oid.
In the end I also tried to load and build the intersection per county and then just concat the results. It’s not faster because of the spatial indexing… The RAM usage is much lower at about 3 GB.
With the reduced number of columns, the running times are:
| Task | Geopandas \s | Geopandas & arrow \s | Geopandas & pyogrio, per county \s |
|---|---|---|---|
| Loading form | 74 | 59 | |
| Intersection | 204 | 181 | |
| Parquet | 11 | 11 | 12 |
| Total | 290 | 251 | 264 |
We have saved 3,620,994 polygons.
Dask-Geopandas
Expectations: Partitioning the DataFrame should increase the number of cores used. This should reduce the computation time.
Observations: I open the shapefiles as before with Geopandas, but then convert them to a Dask-Geopandas GeoDataFrame. All this increases the loading time a bit from about 60s to 76s. It’s not much because I don’t do the spartial partioning!
Finally, I try the map_partitions method. On the left a Dask GeoDataFrame (the larger parcels dataset) and on the right the smaller GeoDataFrame on the right. Having the larger dataset as the Dask-GeoDataFrame increases speed. No, spatial swapping is not necessary as the spatial index is already used. For the map_partitions I create a function that wraps the overlay. This creates a single duplicate.
| Task | Geopandas \s | Dask-Geopandas \s |
|---|---|---|
| Loading form | 59 | 76 |
| Intersection | 181 | 62 |
| Parquet | 11 | 12 |
| Total | 251 | 151 |
This really does use all the cores, and you can see a usage between 30% and 95% while the Overlay is being processed. This reduces the computing time to 33% on this machine.
But three times faster, for 8 cores and 16 threads on the machine. Not quite what I expected.
DuckDB
DuckDB has a spatial extension. Although the csv/parquet file readers work well, the tokens to load multiple files at once. But this is not possible with ST_Read for reading spatial data. So I use pathlib as with the other frameworks. There is no overlay, we have to do all the steps myself. So there is a possibility that my solution is suboptimal.
I was unable to save the data geopackage due to an error in sqlite3_exec, which was unable to open the save tree.
Expectation: Not much, it’s stated to be faster than SQLite for DataAnalysis. Why could test before. But how does it compare to DataFrames, which are also in RAM? Should be faster, due to multicore usage. The memory layout benefits cannot be much, as GeoPandas also uses Apache Arrow?
Observation: CPU usage is high at first, but drops steadily. For Dask the usage fluctuates. I suspect this is due to index usage. The ST_Intersects operation uses the index, ST_Intersection does not.
The execution speed is much slower than for Dask. Saving takes so long that it is even as slow as normal geopandas. Using the database in persistent mode (giving the connection a filename) increases the execution time. Loading takes 70% longer, but we eliminate the need to save the data. Yes, I could load the data into a DataFrame and save it, but then it is no longer a full GeoPandas DuckDB comparison. The saved database is actually a bit smaller than the FlatGeoBuf file. The comparison between DuckDB and Geopandas (with arrow) in speed is
| Task | Geopandas \s | DuckDB (Memory) \s | DuckDB (db-file) \s |
|---|---|---|---|
| Loading Shape | 59 | 67 | 120 |
| Intersection | 181 | 100 | 92 |
| Saving | 11 | 7 | — |
| Overall | 251 | 174 | 212 |
| Polygon Count | 3620994 | 3619038 | — |
DuckDB has a lower count in returned Polyons, but I assume that the missing are in included in the collections. (Not tested.)
Apache Sedona with PySpark
Expectations: Some loss due to virtualization with Docker. So PySpark would not be as fast as Dask?
Although the code is conceptually very similar to the database version. It is an interesting technology. I started with the Sedona container as a docker-compose file. This created a local Spark instance with Sedona and Jupyter notebook.
The shapefiles can be loaded with a placeholder. No iteration (in code) required. But we need to validate the geometry with ST_MakeValid. Otherwise, I get a Java error message which is really long. Which makes it hard to understand, at least if you are not used to it. You can use SQL syntax on the DataFrames, or you can use message chaining methods. I started with SQL code (which is more universal), but it contains long strings in your code. Once everything was working, I switched to method chaining. Which in my eyes looks better, more functional. This flexibility is a plus.
So far the code is lazy. A show method only executes on the row it will show, counting on all rows. Lazy execution can lead to double execution, so I remove all count methods. The slowest part seems to be writing. But differentiating the timing is difficult due to the lazy execution. The data is saved as a geoqarquet file with 3618339 polygons, the size was about 320 MB with snappy compression and 250 MB with ZSTD. Saving as a single parquet file takes about 158 seconds. I would have liked to use more containers on a single device and let them talk to each other to get multiple workers and a master to see how much multi-node reduced the computation time further.
But that did not seem so trivial (please prove me wrong)
Overall speed comparison
For the overall speed and memory usage comparison, I exclude Apache-Sedona as it is running in a Docker container (for now).
The previous timings were based on warmed runs, but single executions. Here we use 10 warmed runs to get a better picture. We need the warming because we use the same input data, so a GeoPandas run would also warm up the Dask GeoPandas run, and so on. Without warming GeoPandas, this would be even slower. These execution times must always be slower than the previous ones, because they include loading the Python interpreter and all libraries. and all libraries.
Without using arrow for input data. GeoPandas took 287.519 s ± 1.532 s to open, overlay and save. The overall variation will be small.
Opening the files with the use_arrow option reduces the computation time by about 8%. The execution takes: 264.590 s ± 4.891 s.
With dask, the speed decreases slightly. The addition of multicore reduces the total execution time by a third. On the other hand, I still have a good part (loading the data) that is limited to single core. So we end up with an execution time of 169.577 s ± 1.882 s.
For DuckDB with in-memory (and saving to FlatGeoBuf) the execution time is 271.455 s ± 2.790 s, and when persisting and not writing the final result it takes 233.427 s ± 6.805 s.
Total Memory Usage Comparison
The question here is whether it is necessary to hold all the data in RAM at the same time, or whether a good strategy can reduce RAM usage. To hold only the final result and partitions of the input data.
The input data accounts for about 8 GB of RAM usage. This is the plateau in RAM usage of the Geopandas program. RAM usage peaks at just under 14 GB.
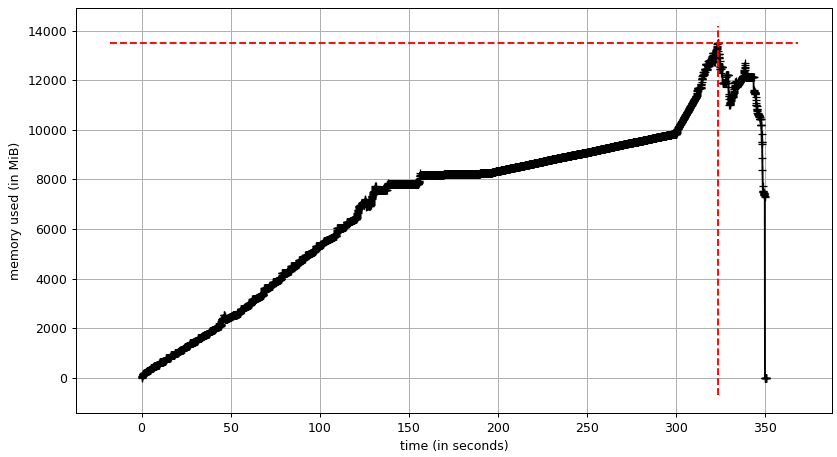
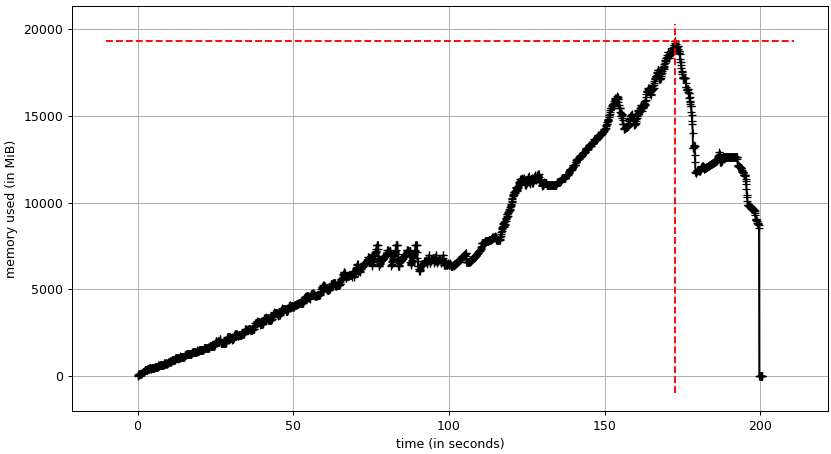
Dask seems to need several copies of the input data. We have a first plateau at about 8 GB and a at about 12 GB. The RAM usage peaks at about 19 GB.
The memory layout in DUCKDB greatly reduces the peak memory usage. I can also use a view for the final result, which adds up to even more savings. Frankly, at only about 7 GB, the peak RAM usage is smaller than the input data in GeoPandas. The input data alone uses about 3.5 GB.
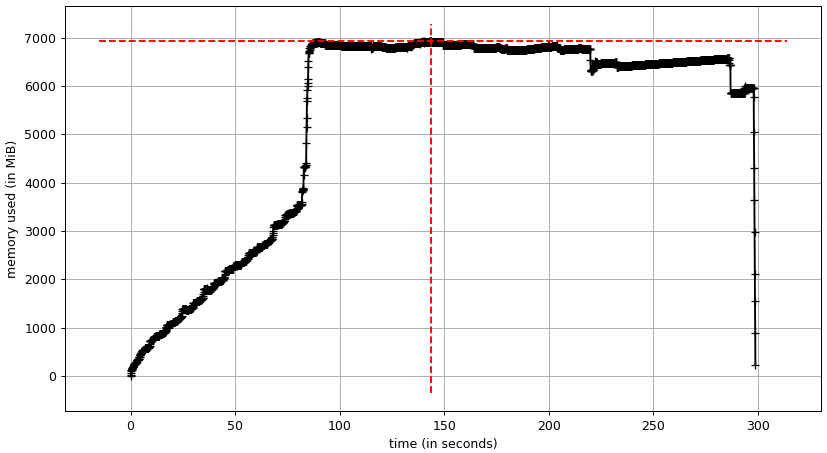
The persistence of the database does not change much, the input data seems to be even smaller at 2.5 GB.
The top RAM usage is also reduced to about 6.2 GB.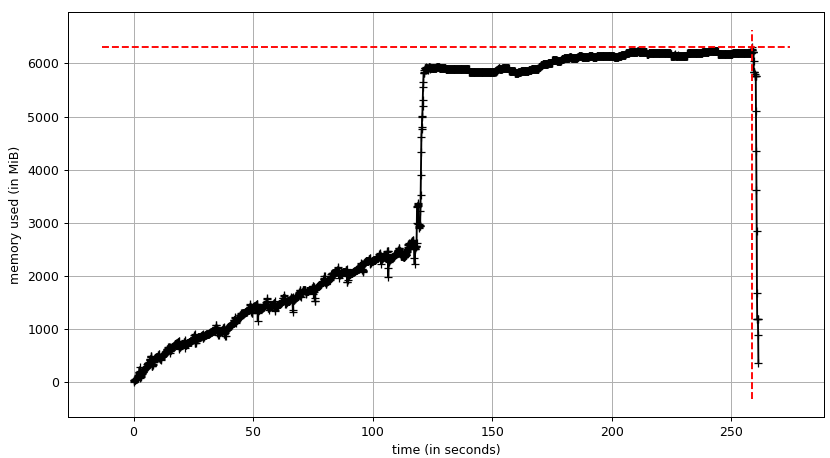
Conclusion
| Package | Total duration \s | Top RAM usage \GB |
|---|---|---|
| Geopandas | 287.5 ± 1.5 | |
| Geopandas (arrow) | 264.6 ± 4.9 | 14 |
| Dask-Geopandas | 169.6 ± 1.9 | 19 |
| DuckDB (in memory) | 167.2 ± 3.0 | 7 |
| DuckDB (persistent) | 233.4 ± 6.8 | 6.2 |
The intersection itself has a speedup S of about three for Dask-GeoDataFrames and two for DuckDB compared to GeoPandas. This is despite using 8 cores with hyper-threading. I suspect that DuckDB is slower here because intersection does not use the spatial index, but intersection does. But DuckDB makes it up with faster loading and saving. When we are able to use multiple cores, loading the data becomes a relatively long part of the total execution time. Either a distribution in geoparquet or loading each file in a separate thread could help.
For Apache-Sedona we can only compare the total execution time and this seems to be on par with Dask-GeoPandas.
If low memory usage is important, DUCKDB is a good option. So either on systems with low memory, or with huge amounts of data. To avoid using swap. Opening shapefiles with DuckDB is slower than with GeoPandas, but that is acceptable, as the exceution is faster. So far The number of supported file formats can be a deal breaker. But I enjoy geoparquet sofar. Remember, DuckDB does not support raster files.
If you already have a Spark cluster, Sedona may be a valid option. So far Dask is the fastest solution, but uses a huge amount of additional memory. Maybe one day I can recommend DuckDB instead.
While using hyperfine on the DuckDB code, I found that a file is created in the temp folder for each output. These files have an uuid part and are never deleted, eg:
buildings_with_parcels_1976_2_temp.fgb. This seems to be a real bug in DuckDB. So while profiling with hyperfine, the temp files are written to disk until the main drive is full. ↩︎
Last modified on 2024-10-19